Publications
Highlights
For a full list see below or go to Google Scholar, PubMed or Research Gate.
For our lab code go to our Kuzmin Lab Code github page.
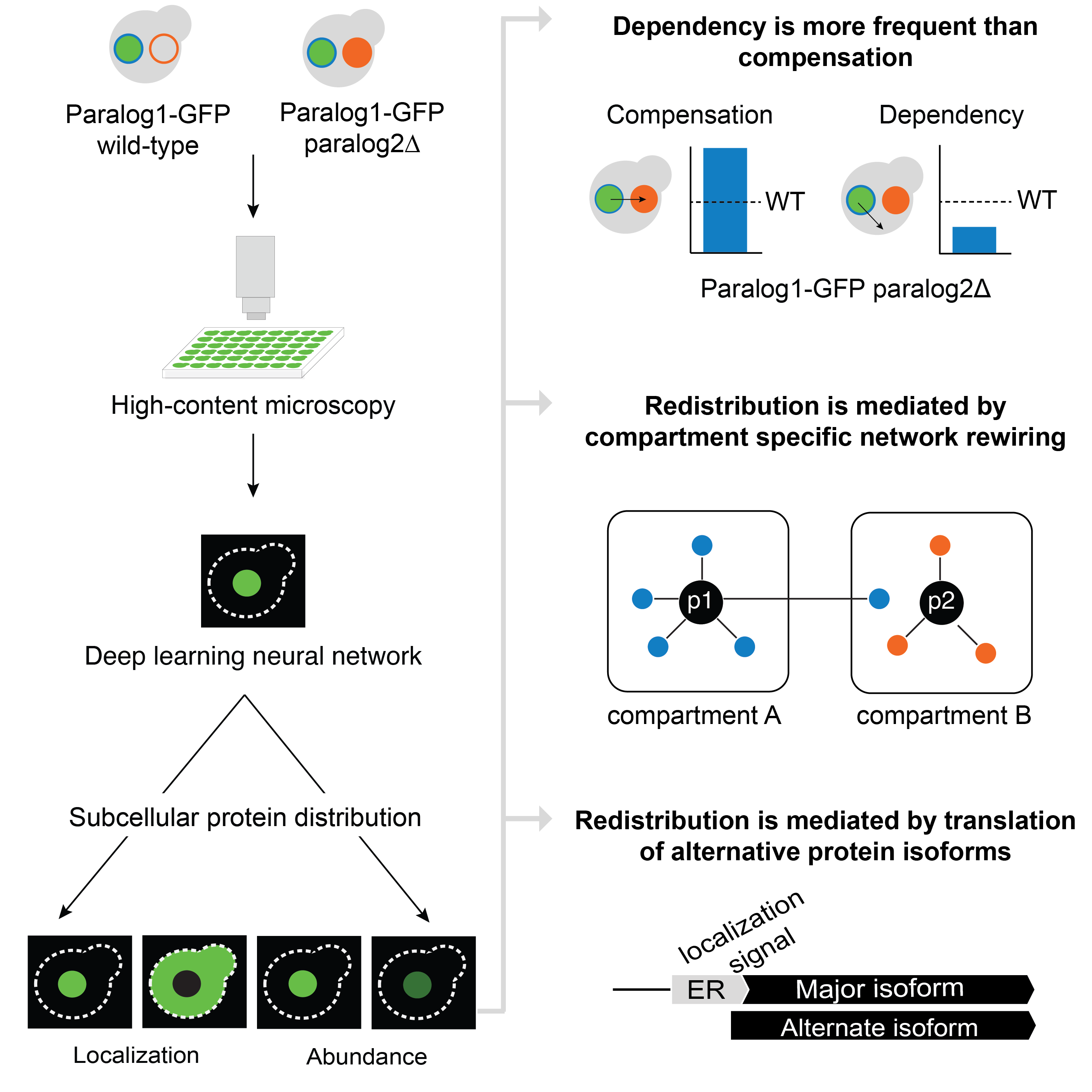
Dandage, R., Papkov, M., Greco, B.M., Pereira, V., Fishman, D., Friesen, H., Wang, K., Styles, E., Kraus, O., Grys, B., Zapata, G., Lefebvre, F., Bourque, G., Boone, C., Andrews, B.J., Parts,L., Kuzmin, E.*
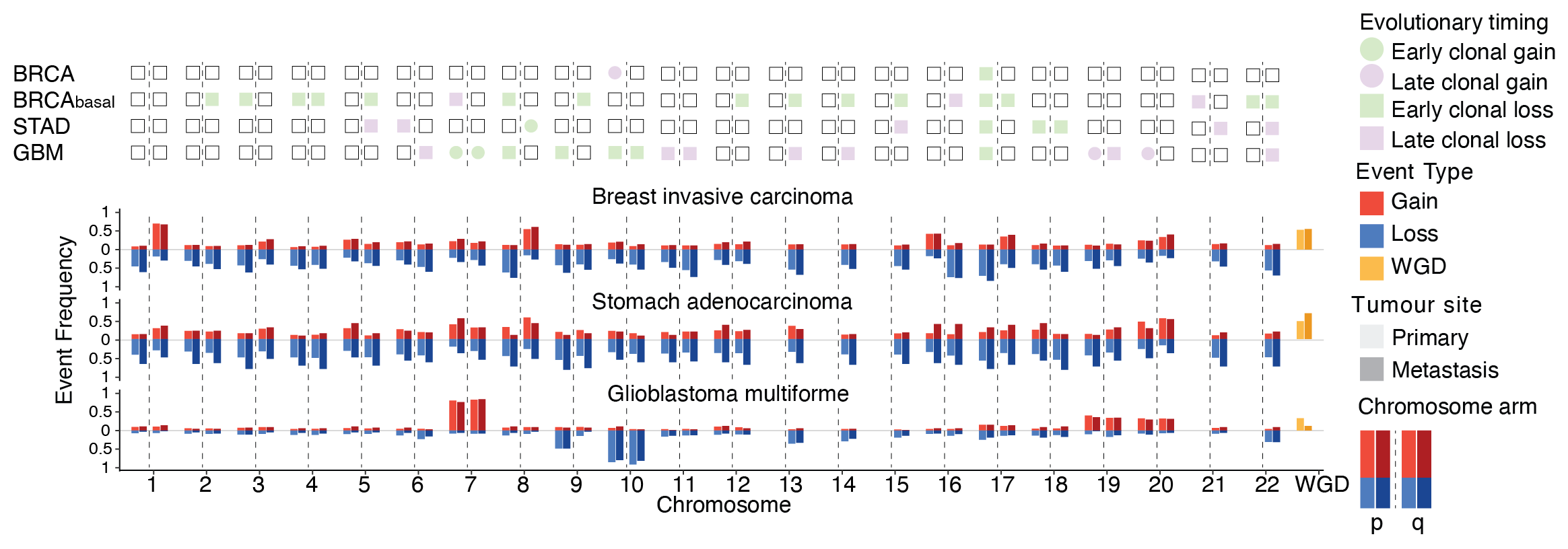
Kuzmin, E., Baker, T.M., Van Loo, P., Glass, L.
Chaos An Interdisciplinary Journal of Nonlinear Science (2024)
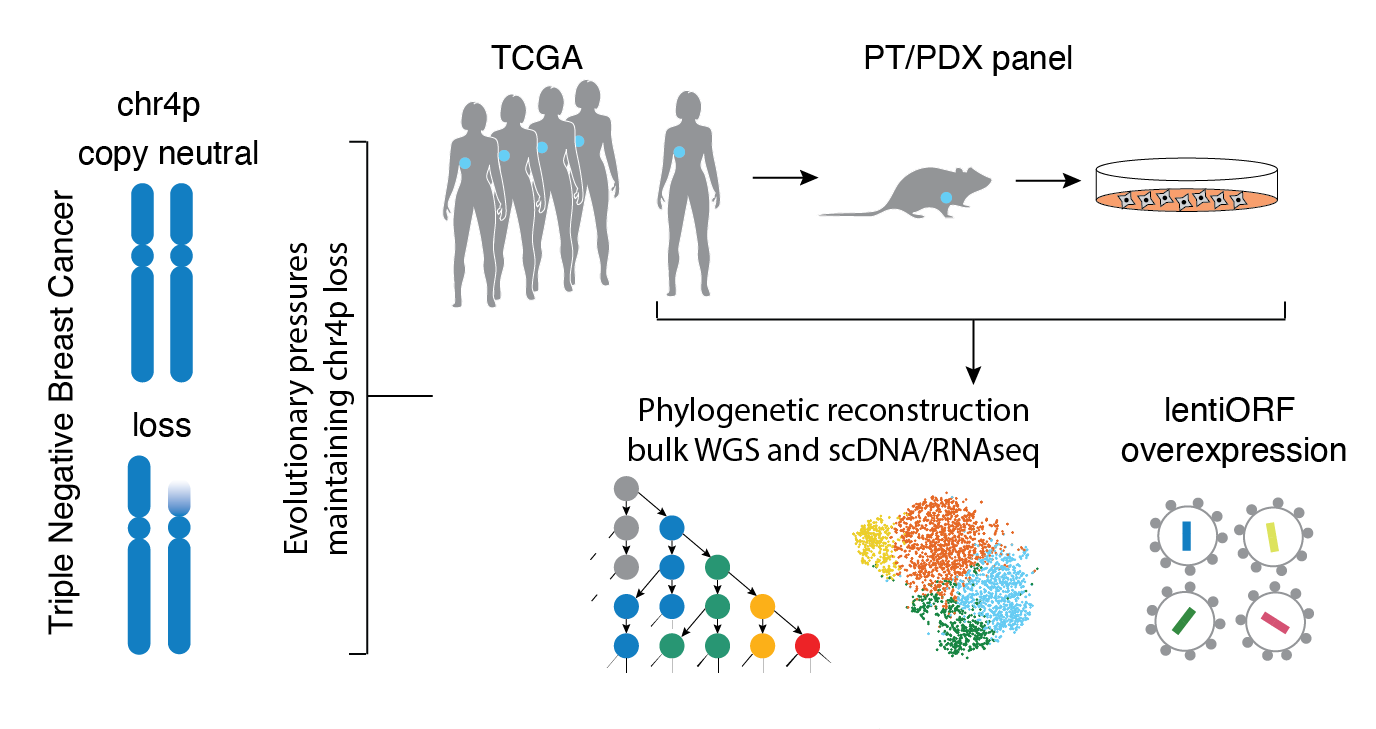
Kuzmin, E., Baker, T.M., Lesluyes, T., Monlong, J., Abe, K.T., Coelho, P.P., Schwartz, M., Zou, D., Morin, G., Pacis, Alain., Yang, Y., Martinez, C., Barber, J., Kuasne, H., Li, R., Bourgey, M., Fortier, A.-M., Davison, P.G., Omeroglu, A., Guiot, M.-C., Morris, Q., Kleinman, C.L., Huang, S., Gingras, A.-C., Ragoussis, J., Bourque, G., Van Loo, P., Park, M.
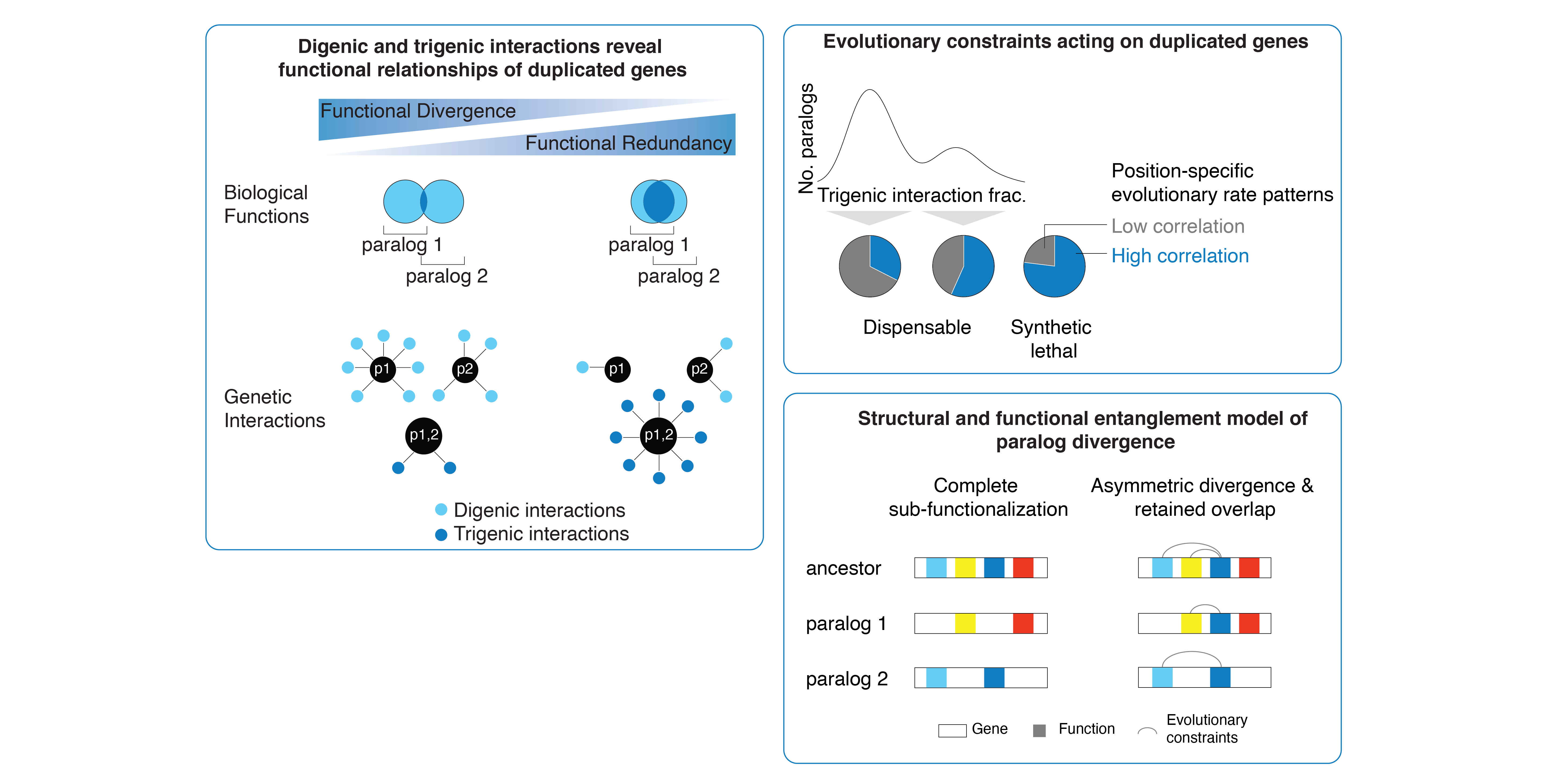
Kuzmin, E., VanderSluis, B., Nguyen Ba, A.N., Wang, W., Koch, N.E., Usaj, M., Khmelinskii, A., Mattiazzi Usaj, M., van Leeuwen, J., Kraus, O., Tresenrider, A., Pryszlak, M., Hu, M.C., Varriano, B., Costanzo, M., Knop, M., Moses, A., Myers, C.L., Andrews, B.J., Boone, C.
Full List
Integrated Metabolic Complex Genetic Interaction Network of Chromosome 4p Loss in Basal Breast Cancer
Karam, L., Pacis, A., Dandage, R., Schwartz, M., Alzial, G., Gherghi, A., Djambazian, H., Zapata, G., Schapfl, M., Asadi, P., Castrillón, M., Orr, A., Hernandez, B., Kargar, P., Vandeloo, M., Santosa, S., Villunger, A., Hart, T., Ragoussis, J., Deblois, G., Bergdahl, A., Park, M., Bourque, G., Kuzmin, E.
bioRxiv (2025)
Quest for orthologs in the era of data deluge and AI: Challenges and innovations in orthology prediction and data integration
Majidian, S., Hadziahmetovic, A., Langschied, F., Pascarelli, S., Prieto-Baños, S., Rojas-Vargas, J., Quest for Orthologs Consortium, Braun, E.L., Dessimoz, C., Durand, D., Fang, G., Gabaldon, T., Glover, N., Liberles, D.A., McWhite, C., Sonnhammer, E.L.L., Ouangraoua, A., Julca, I.
Journal of Molecular Evolution (2025)
PARPAL: PARalog Protein Redistribution using Abundance and Localization in Yeast Database
Greco, B.M., Zapata, G., Dandage, R., Papkov, M., Pereira, V., Lefebvre, F., Bourque, G., Parts,L., Kuzmin, E.*
G3 Genes, Genomes, Genetics (2025)
Single-cell imaging of protein dynamics of paralogs reveals sources of gene retention
Dandage, R., Papkov, M., Greco, B.M., Pereira, V., Fishman, D., Friesen, H., Wang, K., Styles, E., Kraus, O., Grys, B., Zapata, G., Lefebvre, F., Bourque, G., Boone, C., Andrews, B.J., Parts,L., Kuzmin, E.*
iScience (2025)
Benchmarking macaque brain gene expression for horizontal and vertical translation
Luppi, A.I., Liu, Z, Hansen, J.Y., Cofre, R., Kuzmin, E., Froudist-Walsh, S., Palomero-Gallagher, N., Misic, B.
Science Advances (2025)
Dynamics of karyotype evolution
Kuzmin, E., Baker, T.M., Van Loo, P., Glass, L.
Chaos An Interdisciplinary Journal of Nonlinear Science (2024)
Evolution of chromosome arm aberrations in breast cancer through genetic network rewiring
Kuzmin, E., Baker, T.M., Lesluyes, T., Monlong, J., Abe, K.T., Coelho, P.P., Schwartz, M., Zou, D., Morin, G., Pacis, Alain., Yang, Y., Martinez, C., Barber, J., Kuasne, H., Li, R., Bourgey, M., Fortier, A.-M., Davison, P.G., Omeroglu, A., Guiot, M.-C., Morris, Q., Kleinman, C.L., Huang, S., Gingras, A.-C., Ragoussis, J., Bourque, G., Van Loo, P., Park, M.
Cell Reports (2024)
Trans-regulatory variant network contributes to missing heritability
Pereira, V. and Kuzmin, E.*
Cell Genomics (2024)
Correspondence between gene expression and neurotransmitter receptor and transporter density in the human brain
Hansen, J.Y., Markello, R.D., Tuminen, L., Norgaard, M., Kuzmin, E., Palomero-Gallagher, N., Dagher, A., Misic, B.
NeuroImage (2022)
Endosomal LC3C-pathway selectively targets plasma membrane cargo for autophagic degradation
Coelho, P., Hesketh, G., Kuzmin, E., Bell, E., Ratcliffe, C., Gingras, A.C., Park, M.
Nature Communications (2022)
Retention of Duplicated Genes in Evolution
Kuzmin, E., Taylor, J.S., Boone, C.
Trends in Genetics (2022)
Patient-Focused Drug Development: Moving toward better outcomes.
Baca, E., Chatterjee, A., de Boer, D., Kuzmin, E., O’Day, E., Prichep, E.
World Economic Forum (2021)
Inferring Copy Number from Triple Negative Breast Cancer Patient Derived Xenograft scRNAseq data using scCNAutils
Kuzmin, E., Monlong, J., Martinez, C., Kuasne, H., Kleinman, C., Ragoussis, J., Bourque, G., Park, M.
Methods in Molecular Biology (2021)
τ-SGA: synthetic genetic array analysis for systematically screening and quantifying trigenic interactions in yeast
Kuzmin, E., Rahman, M., VanderSluis, B., Costanzo, M., Myers, C.L., Andrews, B.J., Boone, C.
Nature Protocols (2021)
Trigenic Synthetic Genetic Array (τ-SGA) Technique For Complex Interaction Analysis
Kuzmin, E., Andrews, B.J., Boone, C.
Methods in Molecular Biology (2021)
Exploring whole-genome duplicate gene retention with complex genetic interaction analysis
Kuzmin, E., VanderSluis, B., Nguyen Ba, A.N., Wang, W., Koch, N.E., Usaj, M., Khmelinskii, A., Mattiazzi Usaj, M., van Leeuwen, J., Kraus, O., Tresenrider, A., Pryszlak, M., Hu, M.C., Varriano, B., Costanzo, M., Knop, M., Moses, A., Myers, C.L., Andrews, B.J., Boone, C.
Science (2020)
Global Genetic Networks and the Genotype to Phenotype Relationship
Costanzo, M., Kuzmin, E., van Leeuwen, J., Mair, B., Moffat, J., Boone, C., Andrews, B.
Cell (2019)
Dispersible hydrogel force sensors reveal patterns of solid mechanical stress in multicellular spheroid cultures
Lee, W., Kalashnikov, N., Mock, S., Halaoui, R., Kuzmin, E., Putnam A.J., Takayama, S., Park, M., McCaffrey, L., Zhao, R., Leask, R. L., Moraes, C.
Nature Communications (2019)
Systematic Analysis of Complex Genetic Interactions
Kuzmin, E., VanderSluis, B., Wang, W., Tan, G., Deshpande, R., Chen, Y., Usaj, M., Balint, A., Mattiazzi Usaj, M., van Leeuwen, J., Koch, E.N., Pons, C., Dagilis, A.J., Pryszlak, M., Wang, J.Z.Y., Hanchard, J., Riggi, M., Xu, K., Heydari, H., San Luis, B. Shuteriqi, E., Zhu, H., Van Dyk, N., Sharifpoor, S., Costanzo, M., Loewith, R., Caudy, A.A., Bolnick, D., Brown, G.W., Andrews, B.J., Boone, C., Myers, C.L.
Science (2018)
Kibra (WWC1) is a metastatic suppressor gene affected by chromosome 5q loss in human breast cancers
Knight, J.F., Sung, V.Y.C., Kuzmin, E., Couzens, A.L., de Verteuil, D.A., Johnson, R.M., Gruosso, T., Lee, W., Saleh, S.M., Zuo, D., Guiot, M.C., Davis, R.R., Zhao, H., Gregg, J.P., Moraes, C., Gingras, A.C., Park, M.
Cell Reports (2018)
Exploring genetic suppression interactions on a global scale
van Leeuwen, J., Pons, C., Mellor, J.C., Yamaguchi, T.N., Friesen, H., Koschwanez, J., Mattiazzi Ušaj, M., Pechlaner, M., Takar, M., Ušaj, M., VanderSluis, B., Andrusiak, K., Bansal, P., Baryshnikova, A., Boone, C.E., Cao, J., Cote, A., Gebbia, M., Horecka, G., Horecka, I., Kuzmin, E., Legro, N., Liang, W., van Lieshout, N., McNee, M., San Luis, B.J., Shaeri, F., Shuteriqi, E., Sun, S., Yang, L., Youn, J.Y., Yuen, M., Costanzo, M., Gingras, A.C., Aloy, P., Oostenbrink, C., Murray, A., Graham, T.R., Myers, C.L., Andrews, B.J., Roth, F.P., Boone, C.
Science (2016)
A global genetic interaction network maps a wiring diagram of cellular function
Costanzo, M., VanderSluis, B., Koch, E.N., Baryshnikova, A., Pons, C., Tan, G., Wang, W., Usaj, M., Hanchard, J., Lee, S.D., Pelechano, V., Styles, E.B., Billmann, M., van Leeuwen, J., van Dyk, N., Lin, Z., Kuzmin, E., Nelson, J., Piotrowski, J.S., Srikumar, T., Bahr, S., Chen, Y., Deshpande, R., Kurat, C.F., Li, S.C., Li, Z., Mattiazzi Usaj, M., Okada, H., Pascoe, N., San Luis, B.J., Sharifpoor, S., Shuteriqi, E., Simpkins, S.W., Snider, J., Garadi Suresh, H., Tan, Y., Zhu, H., Malod-Dognin, M., Janjic, V., Przulj, N., Troyanskaya, O.G., Stagljar, I., Xia, T., Ohya, Y., Gingras, A.C., Raught, B., Boutros, M., Steinmetz, L.M., Moore, C.L., Rosebrock, A.P., Caudy, A.A., Myers, C.L., Andrews, B., Boone, C.
Science (2016)
Synthetic Genetic Arrays: Automation of Yeast Genetics
Kuzmin, E., Costanzo, M., Andrews, B.J., Boone, C.
Cold Spring Harbor Protocols (2016)
Synthetic Genetic Array Analysis
Kuzmin, E., Costanzo, M., Andrews, B.J., Boone, C.
Cold Spring Harbor Protocols (2016)
Chemical genomics-based antifungal drug discovery: targeting glycosylphosphatidylinositol (GPI) precursor biosynthesis
Mann, P.A., McLellan, C.A., Koseoglu, S., Si, Q., Kuzmin, E., Flattery, A., Harris, G., Sher, X., Murgolo, N., Wang, H., Devito, K., de Pedro, N., Genilloud, O., Nielsen Kahn, J., Jian, B., Costanzo, M., Boone, C., Garlisi, C.G., Lindquist, S., Roemer, T.
ACS Infectious Disease (2015)
Synthetic Genetic Array Analysis for Global Mapping of Genetic Networks in Yeast
Kuzmin, E., Sharifpoor, S., Baryshnikova, A., Costanzo, M., Myers, C.L., Andrews, B., Boone, C.
Methods in Molecular Biology (2014)
A comparative genomic approach for identifying synthetic lethal interactions in human cancer
Deshpande, R., Asiedu, M., Klebig, M., Sutor, S., Kuzmin, E., Nelson, J., Piotrowski, J., Shin, S.H., Yoshida, M., Costanzo, M., Boone, C., Wigle, D.A., Myers, C.L.
Cancer Research (2013)
SGAtools: one-stop analysis and visualization of array-based genetic interaction screens
Wagih, O., Usaj, M., Baryshnikova, A., Vandersluis, B., Kuzmin, E., Costanzo, M., Myers, C.L., Andrews, B.J., Boone, C.M., Parts, L.
Nucleic Acids Research (2013)
Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell
Magtanong, L., Ho, C.H., Barker, S.L.*, Jiao, W., Baryshnikova, A., Bahr, S., Smith, A.M., Heisler, L.E., Choy, J.S., Kuzmin, E., Andrusiak, K., Kobylianski, A., Li, Z., Costanzo, M., Basrai, M.A., Giaever, G., Nislow, C., Andrews, B., Boone, C.
Nature Biotechnology (2011)
Transient limb ischemia remotely preconditions through a humoral mechanism acting directly on the myocardium: evidence suggesting cross-species protection
Shimizu, M., Tropak, M., Diaz, R.J., Suto, F., Surendra, H., Kuzmin, E., Li, J., Gross, G., Wilson, G.J., Callahan, J.W., Redington, A.N.
Clinical Science (2009)