Research
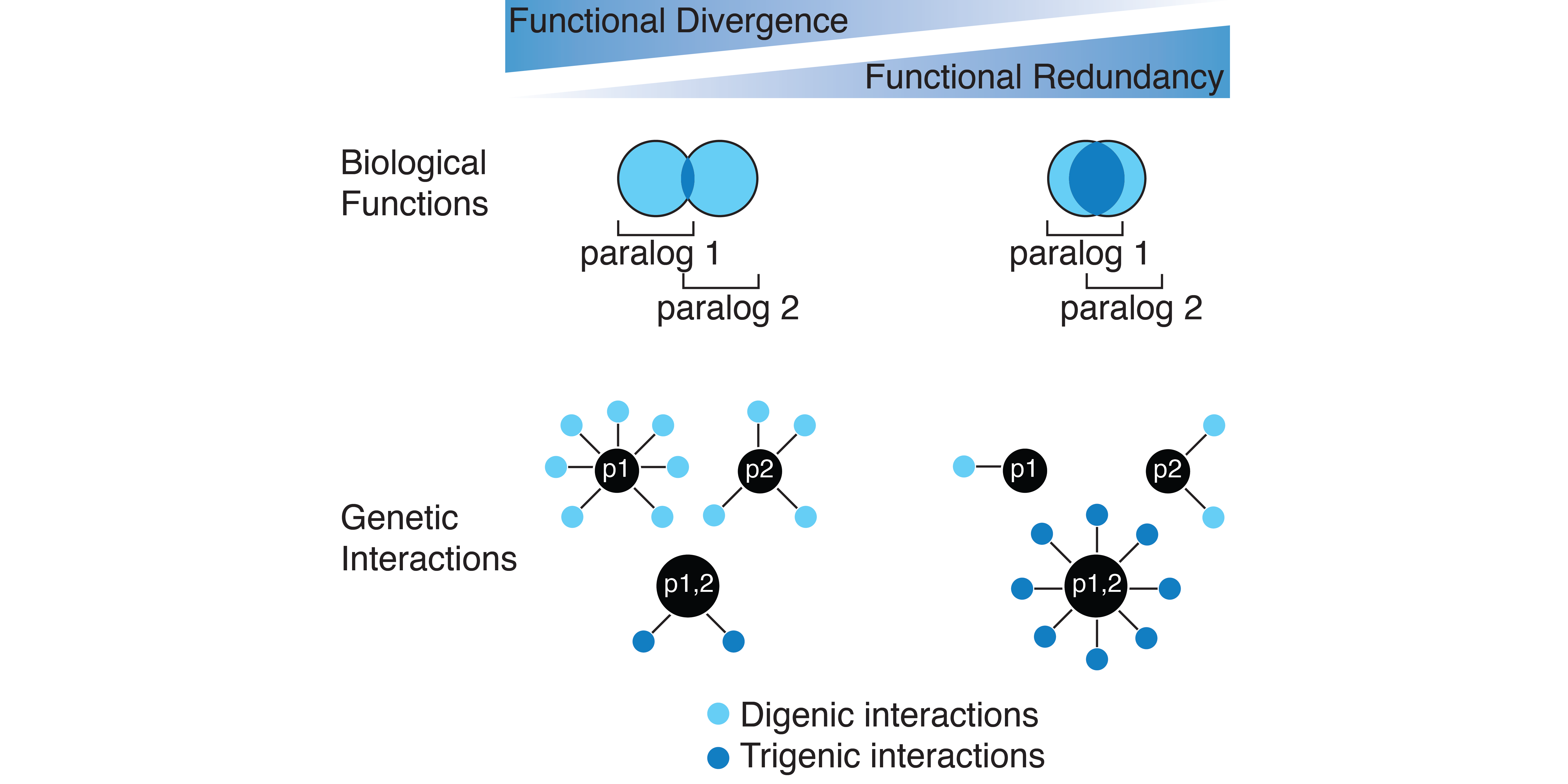
A central question in evolution has remained the persistence of duplicated genes over millions of years. We will use trigenic synthetic genetic array methodology in yeast to investigate the functional redundancy and divergence of duplicated genes using complex genetic interactions. Whole genome duplication in yeast co-incided with the evolution of flesh fruiting plants (angiosperms). Changes to the metabolic niche of yeast likely exerted selective pressures and retention of a set of duplicated genes.
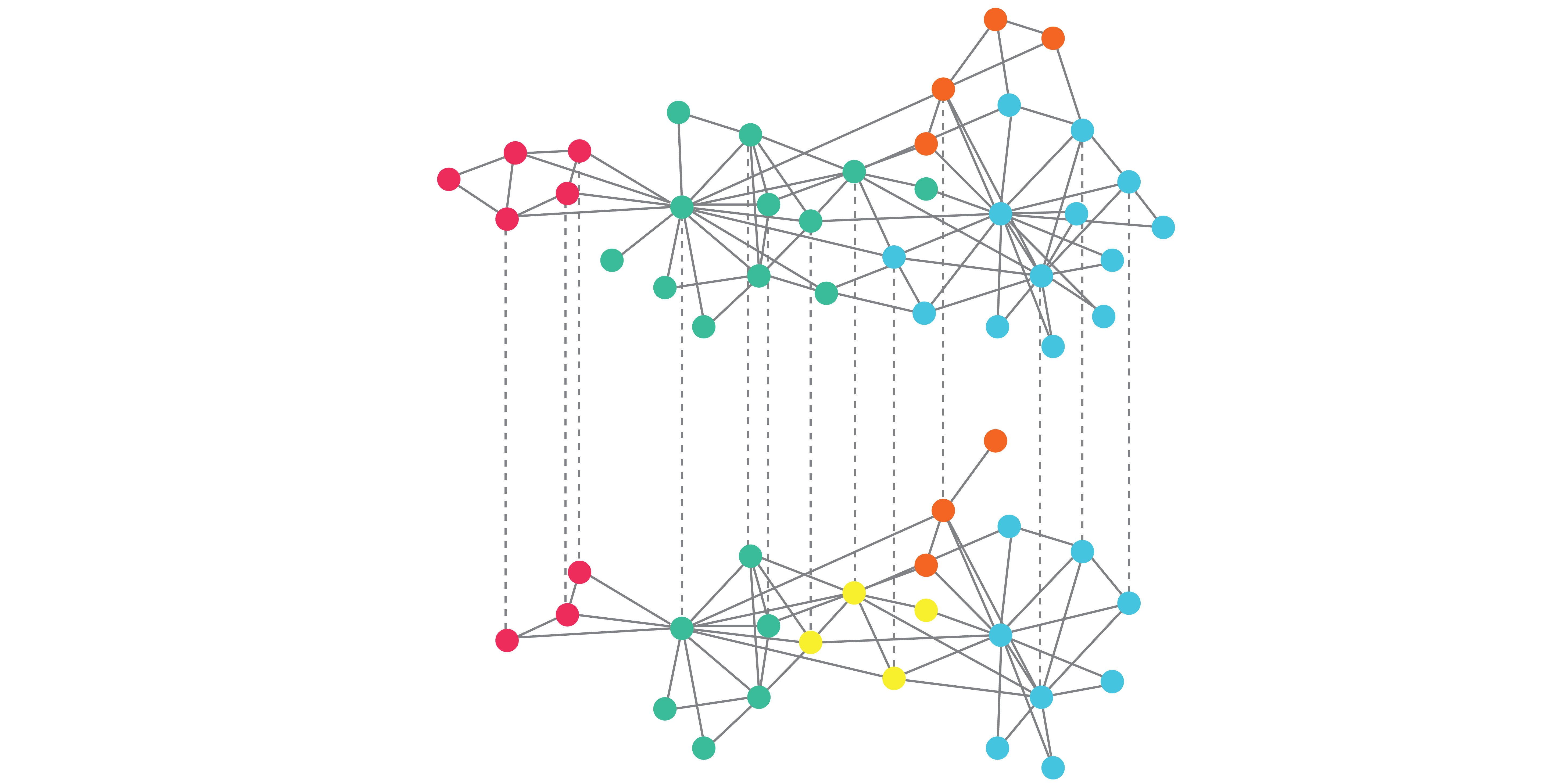
An outstanding question in the field, is the extent of genetic interaction network conservation. We will use trigenic synthetic genetic array methodology in yeast to interrogate complex genetic interaction network rewiring between distantly related yeast species and integrate the findings with other species. This approach will enable us to determine whether genetic network rewiring involving complex genetic interactions can expand the level of conservation of genetic interactions between evolutionary distant organisms.
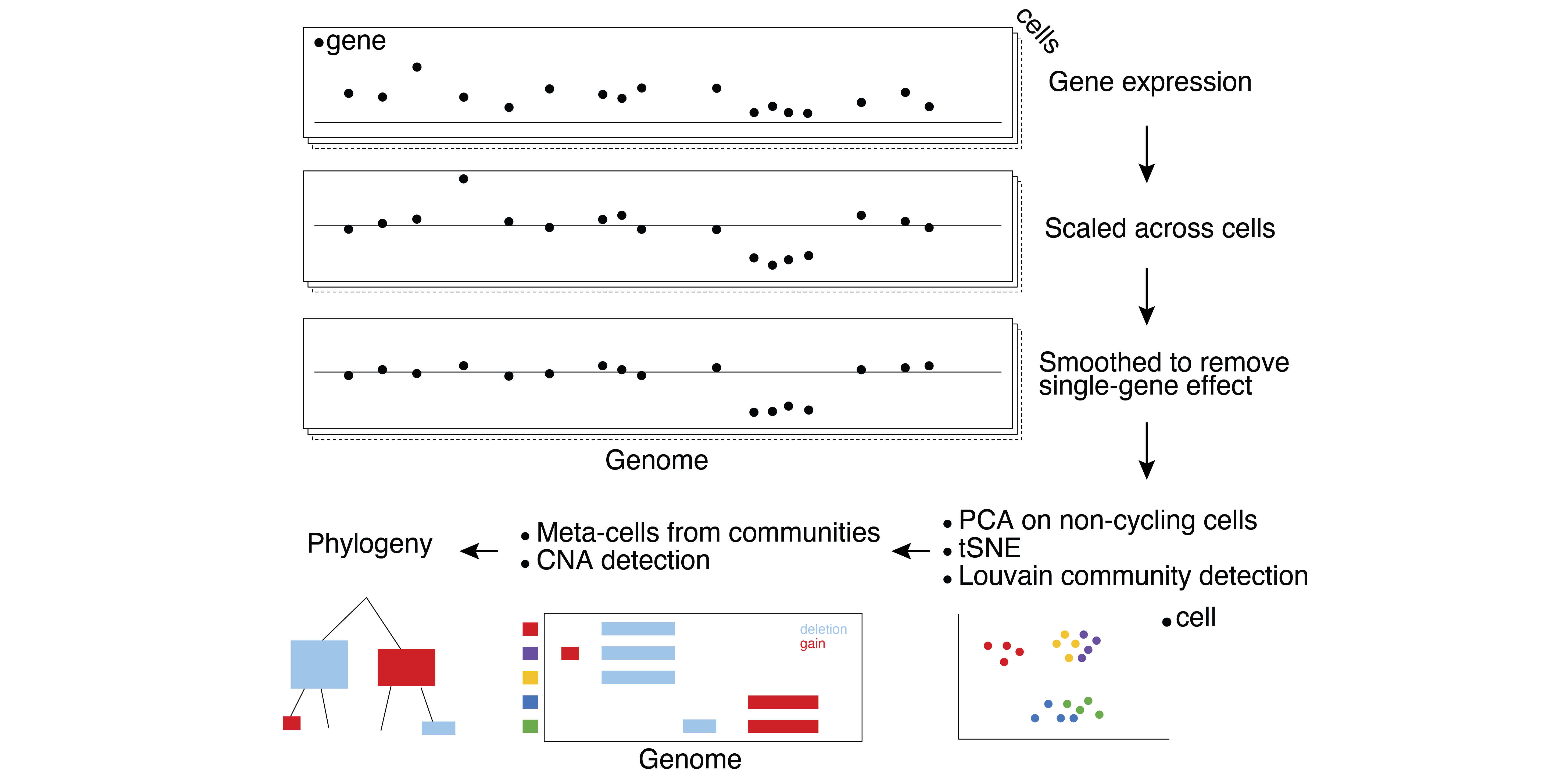
We previously showed that large chromosomal deletions are maintained in triple negative breast cancer due to evolutionary early genetic network rewiring rendering multiple genes within this region to be dosage sensitive. It remains to be uncovered what is the extent of genetic interactions involving tumor suppressors within and between large chromosomal deletions. We will use a combination of pooled and arrayed CRISPR screens with a cell proliferation and gene expression readout to address this question.
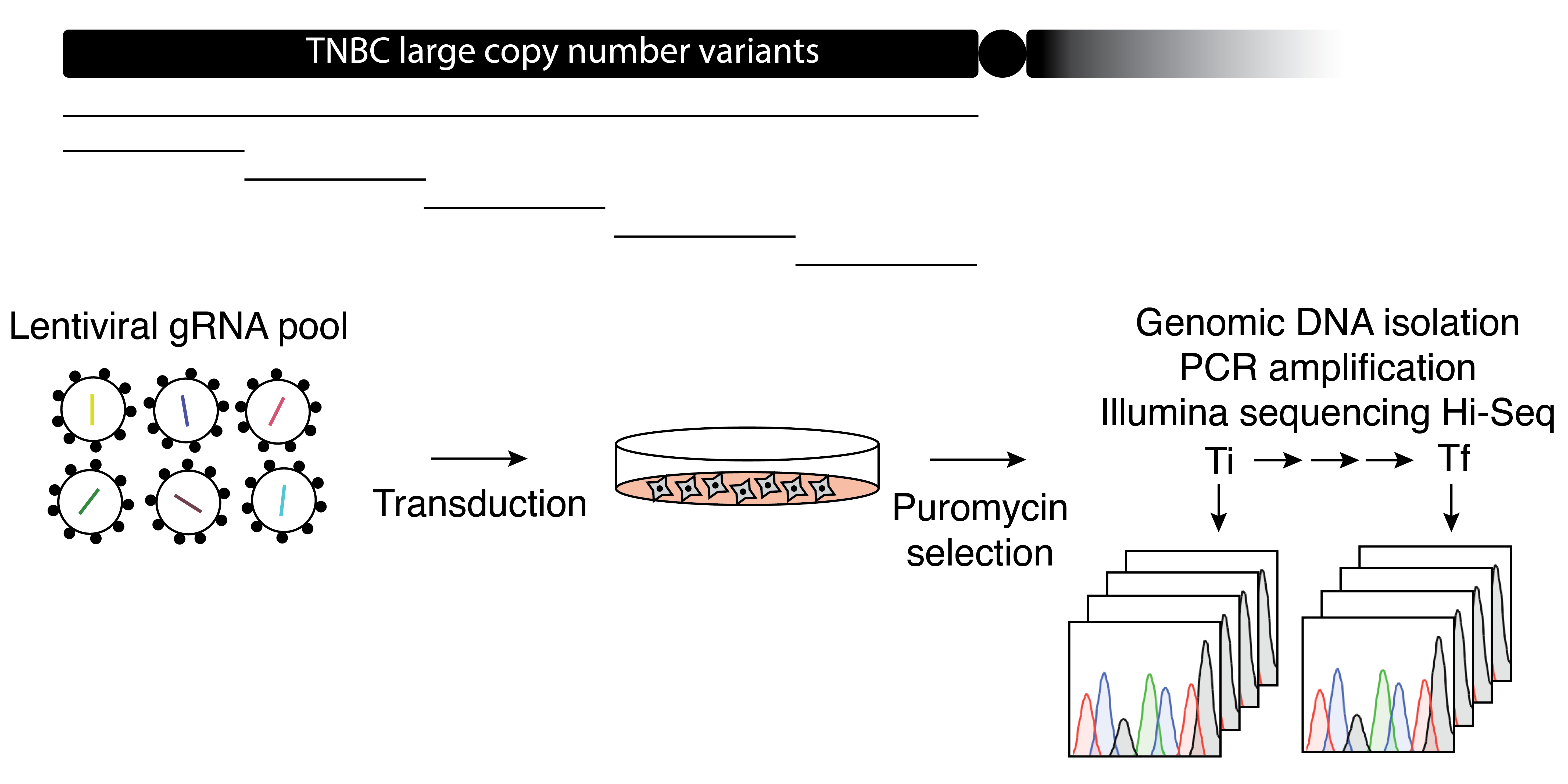
Large chromosomal alterations are very common in cancer yet an understanding of their therapeutic potential for precision medicine is lacking. We will investigate whether large chromosomal deletions represent genetic vulnerabilities, which are therapeutically relevant in cancer. To accomplish this goal we will use pooled and arrayed CRISPR screens in preclinical models of cancer, such as triple negative breast cancer patient-derived xenograft cell lines and organoids.